Partial Dependence for 2 Features Explainer Demo
This example demonstrates how to interpret a model using the H2O Eval Studio library and retrieve the data and partial dependence plot for 2 featues.
[1]:
import logging
import os
import daimojo
import datatable
import webbrowser
from h2o_sonar import interpret
from h2o_sonar.lib.api import commons, explainers
from h2o_sonar.lib.api.models import ModelApi
from h2o_sonar.explainers.pd_2_features_explainer import PdFor2FeaturesExplainer
[2]:
# dataset
dataset_path = "../../data/creditcard.csv"
target_col = "default payment next month"
# model
mojo_path = "../../data/models/creditcard-binomial.mojo"
mojo_model = daimojo.model(mojo_path)
model = ModelApi().create_model(
model_src=mojo_model,
target_col=target_col,
used_features=list(mojo_model.feature_names),
)
# results
results_location = "./results"
os.makedirs(results_location, exist_ok=True)
[3]:
# explainer description
interpret.describe_explainer(PdFor2FeaturesExplainer)
[3]:
{'id': 'h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer',
'name': 'PdFor2FeaturesExplainer',
'display_name': 'Partial Dependence Plot for Two Features',
'description': 'Partial dependence for 2 features portrays the average\nprediction behavior of a model across the domains of two input variables\ni.e. interaction of feature tuples with the prediction. While PD for one feature\nproduces 2D plot, PD for two features produces 3D plots. This explainer plots PD for\ntwo features using heatmap, contour 3D or surface 3D.\n',
'model_types': ['iid', 'time_series'],
'can_explain': ['regression', 'binomial'],
'explanation_scopes': ['global_scope'],
'explanations': [{'explanation_type': 'global-partial-dependence',
'name': 'PartialDependenceExplanation',
'category': None,
'scope': 'global',
'has_local': None,
'formats': []}],
'parameters': [{'name': 'sample_size',
'description': 'Sample size for Partial Dependence Plot of 2 features.',
'comment': '',
'type': 'int',
'val': 25000,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'max_features',
'description': 'Partial Dependence Plot number of features.',
'comment': '',
'type': 'int',
'val': 3,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'features',
'description': 'List of features from which to choose pairs to compute PD for two features.',
'comment': '',
'type': 'list',
'val': None,
'predefined': [],
'tags': ['SOURCE_DATASET_COLUMN_NAMES'],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'grid_resolution',
'description': 'Partial Dependence Plot observations per bin (number of equally spaced points used to create bins).',
'comment': '',
'type': 'int',
'val': 10,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'oor_grid_resolution',
'description': 'Partial Dependence Plot number of out of range bins.',
'comment': '',
'type': 'int',
'val': 0,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'quantile-bin-grid-resolution',
'description': 'Partial Dependence Plot quantile binning (total quantile points used to create bins).',
'comment': '',
'type': 'int',
'val': 0,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'plot_type',
'description': 'Plot type.',
'comment': '',
'type': 'str',
'val': 'heatmap',
'predefined': ['heatmap', 'contour-3d', 'surface-3d'],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''}],
'keywords': []}
Interpretation
[4]:
interpretation = interpret.run_interpretation(
dataset=dataset_path,
model=model,
target_col=target_col,
results_location=results_location,
log_level=logging.INFO,
explainers=[
commons.ExplainerToRun(
explainer_id=PdFor2FeaturesExplainer.explainer_id(),
params="",
)
]
)
/home/srasaratnam/projects/h2o-sonar/venv/lib/python3.8/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 10.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 10.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 10.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 10.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 20.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 30.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 40.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 50.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 60.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 70.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 70.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 70.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 70.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 70.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 70.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 80.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 80.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 80.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 80.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 80.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 90.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 90.0%
h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer: progress 90.0%
FixedFormatter should only be used together with FixedLocator
FixedFormatter should only be used together with FixedLocator
FixedFormatter should only be used together with FixedLocator
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
Explainer Result
[5]:
# retrieve the result
result = interpretation.get_explainer_result(PdFor2FeaturesExplainer.explainer_id())
[6]:
# open interpretation HTML report in web browser
webbrowser.open(interpretation.result.get_html_report_location())
[6]:
True
[7]:
# summary
result.summary()
[7]:
{'id': 'h2o_sonar.explainers.pd_2_features_explainer.PdFor2FeaturesExplainer',
'name': 'PdFor2FeaturesExplainer',
'display_name': 'Partial Dependence Plot for Two Features',
'description': 'Partial dependence for 2 features portrays the average\nprediction behavior of a model across the domains of two input variables\ni.e. interaction of feature tuples with the prediction. While PD for one feature\nproduces 2D plot, PD for two features produces 3D plots. This explainer plots PD for\ntwo features using heatmap, contour 3D or surface 3D.\n',
'model_types': ['iid', 'time_series'],
'can_explain': ['regression', 'binomial'],
'explanation_scopes': ['global_scope'],
'explanations': [{'explanation_type': 'global-report',
'name': 'Partial Dependence Plot for Two Features',
'category': 'DAI MODEL',
'scope': 'global',
'has_local': None,
'formats': ['text/markdown']},
{'explanation_type': 'global-3d-data',
'name': 'Partial Dependence Plot for Two Features',
'category': 'DAI MODEL',
'scope': 'global',
'has_local': None,
'formats': ['application/json', 'application/vnd.h2oai.json+csv']},
{'explanation_type': 'global-html-fragment',
'name': 'Partial Dependence Plot for Two Features',
'category': 'DAI MODEL',
'scope': 'global',
'has_local': None,
'formats': ['text/html']}],
'parameters': [{'name': 'sample_size',
'description': 'Sample size for Partial Dependence Plot of 2 features.',
'comment': '',
'type': 'int',
'val': 25000,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'max_features',
'description': 'Partial Dependence Plot number of features.',
'comment': '',
'type': 'int',
'val': 3,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'features',
'description': 'List of features from which to choose pairs to compute PD for two features.',
'comment': '',
'type': 'list',
'val': None,
'predefined': [],
'tags': ['SOURCE_DATASET_COLUMN_NAMES'],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'grid_resolution',
'description': 'Partial Dependence Plot observations per bin (number of equally spaced points used to create bins).',
'comment': '',
'type': 'int',
'val': 10,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'oor_grid_resolution',
'description': 'Partial Dependence Plot number of out of range bins.',
'comment': '',
'type': 'int',
'val': 0,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'quantile-bin-grid-resolution',
'description': 'Partial Dependence Plot quantile binning (total quantile points used to create bins).',
'comment': '',
'type': 'int',
'val': 0,
'predefined': [],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''},
{'name': 'plot_type',
'description': 'Plot type.',
'comment': '',
'type': 'str',
'val': 'heatmap',
'predefined': ['heatmap', 'contour-3d', 'surface-3d'],
'tags': [],
'min_': 0.0,
'max_': 0.0,
'category': ''}],
'keywords': []}
[8]:
# parameters
result.params()
[8]:
{'sample_size': 25000,
'max_features': 3,
'features': None,
'grid_resolution': 10,
'oor_grid_resolution': 0,
'quantile-bin-grid-resolution': 0,
'plot_type': 'heatmap'}
Display PD Data
[9]:
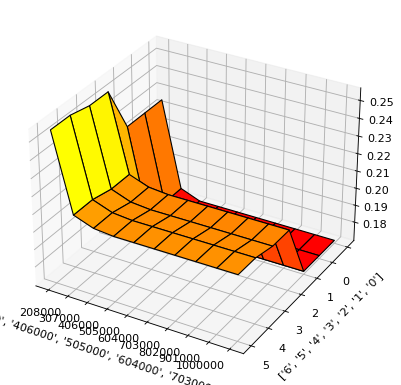
result.data(feature_names="'LIMIT_BAL' and 'EDUCATION'")
[9]:
{'10000': {'6': 0.2552240192890167,
'5': 0.2552240192890167,
'4': 0.25266972184181213,
'3': 0.25266972184181213,
'2': 0.2251938432455063,
'1': 0.2251938432455063,
'0': 0.2251938432455063},
'208000': {'6': 0.21158096194267273,
'5': 0.21158096194267273,
'4': 0.20898468792438507,
'3': 0.20898468792438507,
'2': 0.17519091069698334,
'1': 0.17519091069698334,
'0': 0.17519091069698334},
'307000': {'6': 0.20750007033348083,
'5': 0.20750007033348083,
'4': 0.20484082400798798,
'3': 0.20484082400798798,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'406000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'505000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'604000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'703000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'802000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'901000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279},
'1000000': {'6': 0.20645806193351746,
'5': 0.20645806193351746,
'4': 0.2037988305091858,
'3': 0.2037988305091858,
'2': 0.17081709206104279,
'1': 0.17081709206104279,
'0': 0.17081709206104279}}
Plot PD Data
[10]:
result.plot(feature_names="'LIMIT_BAL' and 'EDUCATION'")
FixedFormatter should only be used together with FixedLocator
Save Explainer Log and Data
[11]:
# save the explainer log
result.log(path="./pd-2-features-demo.log")
[12]:
!head pd-ice-demo.log
2023-03-12 23:27:53,358 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 BEGIN calculation
2023-03-12 23:27:53,359 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 loading dataset
2023-03-12 23:27:53,360 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 loaded dataset has 10000 rows and 25 columns
2023-03-12 23:27:53,360 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 getting features list, importanceand metadata
2023-03-12 23:27:53,361 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 all most important model features: ['ID', 'LIMIT_BAL', 'SEX', 'EDUCATION', 'MARRIAGE', 'AGE', 'PAY_0', 'PAY_2', 'PAY_3', 'PAY_4', 'PAY_5', 'PAY_6', 'BILL_AMT1', 'BILL_AMT2', 'BILL_AMT3', 'BILL_AMT4', 'BILL_AMT5', 'BILL_AMT6', 'PAY_AMT1', 'PAY_AMT2', 'PAY_AMT3', 'PAY_AMT4', 'PAY_AMT5', 'PAY_AMT6']
2023-03-12 23:27:53,362 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 features used by model: ['ID', 'LIMIT_BAL', 'SEX', 'EDUCATION', 'MARRIAGE', 'AGE', 'PAY_0', 'PAY_2', 'PAY_3', 'PAY_4', 'PAY_5', 'PAY_6', 'BILL_AMT1', 'BILL_AMT2', 'BILL_AMT3', 'BILL_AMT4', 'BILL_AMT5', 'BILL_AMT6', 'PAY_AMT1', 'PAY_AMT2', 'PAY_AMT3', 'PAY_AMT4', 'PAY_AMT5', 'PAY_AMT6']
2023-03-12 23:27:53,362 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885: calculating PD for features ['ID', 'LIMIT_BAL', 'SEX', 'EDUCATION', 'MARRIAGE', 'AGE', 'PAY_0', 'PAY_2', 'PAY_3', 'PAY_4']
2023-03-12 23:27:53,363 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 feature metadata: {'id': [], 'categorical': [], 'numeric': [], 'catnum': [], 'date': [], 'time': [], 'datetime': [], 'text': [], 'image': [], 'date-format': [], 'quantile-bin': {}}
2023-03-12 23:27:53,363 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 1 frame strategy: True
2023-03-12 23:27:53,364 INFO PD/ICE a30a4f57-9c78-4c5f-9c8e-85198b523c1c/745668cc-bbc8-42be-9b39-47487112a885 residual PD/ICE should NOT be calculated, but y has been specified - setting it None
[13]:
# save the explainer data
result.zip(file_path="./pd-ice-demo-archive.zip")
[14]:
!unzip -l pd-ice-demo-archive.zip
Archive: pd-ice-demo-archive.zip
Length Date Time Name
--------- ---------- ----- ----
4474 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/result_descriptor.json
122 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_report/text_markdown.meta
2642 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_report/text_markdown/explanation.md
92249 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_report/text_markdown/image-1.png
80252 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_report/text_markdown/image-2.png
75679 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_report/text_markdown/image-0.png
92249 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/work/image-1.png
80252 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/work/image-2.png
75679 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/work/image-0.png
1247 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/work/explanation.html
40216 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/work/mli_dataset_y_hat.jay
2642 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/work/report.md
110 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_html_fragment/text_html.meta
92249 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_html_fragment/text_html/image-1.png
80252 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_html_fragment/text_html/image-2.png
75679 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_html_fragment/text_html/image-0.png
1247 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_html_fragment/text_html/explanation.html
141 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_json.meta
161 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_vnd_h2oai_json_csv.meta
2583 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_json/data3d_feature_1_class_0.json
1484 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_json/explanation.json
902 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_json/data3d_feature_0_class_0.json
498 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_json/data3d_feature_2_class_0.json
285 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_vnd_h2oai_json_csv/data3d_feature_2_class_0.csv
1466 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_vnd_h2oai_json_csv/data3d_feature_1_class_0.csv
1481 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_vnd_h2oai_json_csv/explanation.json
475 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/global_3d_data/application_vnd_h2oai_json_csv/data3d_feature_0_class_0.csv
2 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/model_problems/problems_and_actions.json
2054 2023-03-12 23:35 explainer_h2o_sonar_explainers_pd_2_features_explainer_PdFor2FeaturesExplainer_993771ed-6505-450c-b5dd-4d81f871d218/log/explainer_run_993771ed-6505-450c-b5dd-4d81f871d218.log
--------- -------
808772 29 files
[ ]: